Downloading Data and Dandisets¶
You can download the content of a Dandiset using the DANDI Web application (such a specific file) or entire Dandisets using the DANDI Python CLI.
Using the DANDI Web Application¶
Once you have the Dandiset you are interested in (see more in the Dandiset View section), you can download the content of the Dandiset.
On the landing page of each Dandiset, you can find Download button on the right-hand panel. After clicking the
button, you will see the specific command you can use with DANDI Python CLI (as well as the information on how to download the CLI).
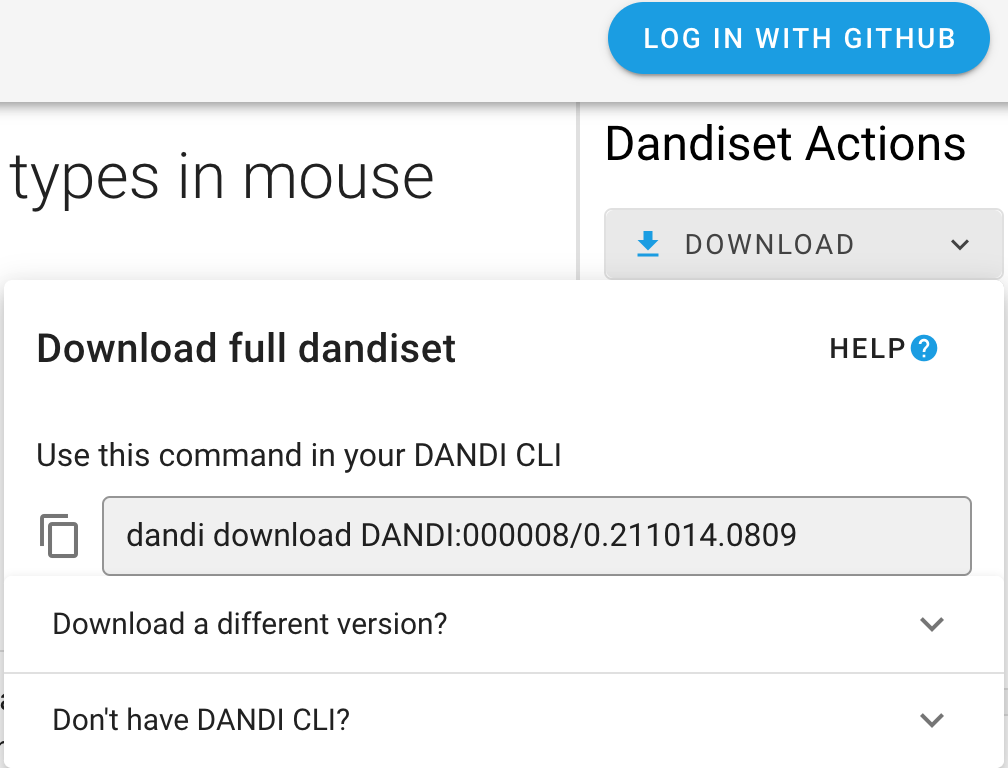
Download specific files¶
The right-side panel of the Dandiset landing page allows you also to access the list of folders and files.
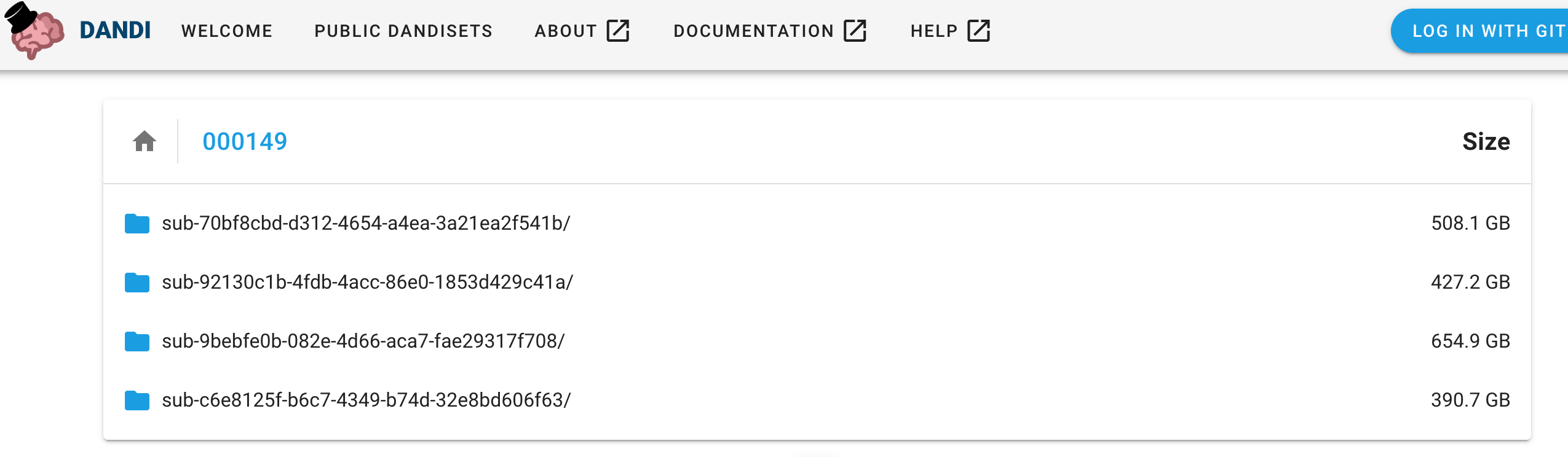
Each file in the Dandiset has a download icon next to it, clicking the icon will start the download process.
Using the Python CLI Client¶
The DANDI Python client gives you more options, such as downloading entire Dandisets.
Before You Begin: You need to have Python 3.8+ and install the DANDI Python Client using pip install dandi.
If you have an issue using the Python CLI, see the Dandi Debugging section.
Download a Dandiset¶
To download an entire Dandiset, you can use the same command as suggested by DANDI web application, e.g.:
dandi download DANDI:000023
Download data for a specific subject from a Dandiset¶
You can download data for specific subjects.
Names of the subjects can be found on DANDI web application or by running a command with the DANDI CLI: dandi ls -r
DANDI:000023.
Once you have the subject ID, you can download the data, e.g.:
dandi download https://api.dandiarchive.org/api/dandisets/000023/versions/draft/assets/?path=sub-811677083
You could replace draft with a specific non-draft version you are interested in (e.g. 0.210914.1900 in the case of this Dandiset), if you are not interested in the latest, possibly different state of the Dandiset.
You can also use the link from DANDI web application, e.g.:
dandi download https://dandiarchive.org/dandiset/000023/0.210914.1900/files?location=sub-541516760%2F
Download a specific file from a Dandiset¶
You can download a specific file from a Dandiset when the link for the specific file can be found on the DANDI web application, e.g.:
dandi download https://api.dandiarchive.org/api/dandisets/000023/versions/0.210914.1900/assets/1a93dc97-327d-4f9c-992d-c2149e7810ae/download/
Hint: dandi download supports a number of Resource Identifiers to point to a Dandiset, folder, or file. Providing
an incorrect URL (e.g. dandi download wrongurl) will provide a list of supported identifiers.
Download the dandiset.yaml file and a specific file within the directory tree of the Dandiset¶
Now available in version 0.63.0 is the --preserve-tree option.
In the command below, replace the <dandiset-id>, <version>, and asset <path>.
The <path> can be found by selecting the View asset metadata icon next to an asset on https://dandiarchive.org and locating the path key.
dandi download --preserve-tree dandi://dandi/<dandiset-id>@<version>/<path>
For example:
dandi download --preserve-tree dandi://dandi/000026@draft/sub-I58/ses-Hip-CT/micr/sub-I58_sample-01_chunk-01_hipCT.json
Using DataLad¶
All dandisets are regularly mirrored to DataLad datasets which are made available at the GitHub organization https://github.com/dandisets. Where present, individual Zarr files are included as subdatasets (git submodules) hosted in the GitHub organization https://github.com/dandizarrs/.
The Git revision histories of each dataset reflect the Dandiset's draft state as of each execution of the mirroring job. Published Dandiset versions are tagged with Git tags.
With DataLad, you can: - clone an entire dataset, - use a specific version of it, - explore history of modifications, - download content of files of interest, - locally discard the content of no-longer-needed files, - use the dataset in a reproducible manner, - include it as a subdataset in your own DataLad dataset, - use https://github.com/datalad/datalad-fuse/ to FUSE-mount individual locally-cloned dandisets so that their files' contents are transparently streamed to your DANDI/DataLad-unaware tools, - etc.
Learn more about DataLad from its handbook at https://handbook.datalad.org/.
Developers' note: DataLad datasets are created using the dandi/backups2datalad tool which is also available for use by the community to similarly maintain mirrors of independent DANDI deployments as DataLad datasets.
Using WebDAV¶
DANDI provides a WebDAV service at https://webdav.dandiarchive.org/ for accessing the data in the DANDI archive. You can use any WebDAV client or even a web browser to access the data - any dandiset, any version, any file or collection of files. You can use any web download tool to download the data from the DANDI archive, e.g.
wget -r -np -nH --cut-dirs=3 https://webdav.dandiarchive.org/dandisets/000027/releases/0.210831.2033/
for a download of a specific release 0.210831.2033 of the 000027 dandiset.
Note: The WebDAV service does not directly serve any file contents; it instead relies on redirects to AWS S3 storage where the contents are stored.
You might need to configure your WebDAV client to follow redirects; e.g., for the davfs2 WebDAV client, set follow_redirect to 1 in /etc/davfs2/davfs2.conf.
Developers' note: The WebDAV service's code is available at https://github.com/dandi/dandidav/ and can also be used for independent DANDI deployments.